Inaccurate results from microbenchmark
2014-02-02 at 2:31 pm 8 comments
The microbenchmark package is a popular way of comparing the time it takes to evaluate different R expressions — perhaps more popular than the alternative of just using system.time to see how long it takes to execute a loop that evaluates an expression many times. Unfortunately, when used in the usual way, microbenchmark can give inaccurate results.
The inaccuracy of microbenchmark has two main sources — first, it does not correctly allocate the time for garbage collection to the expression that is responsible for it, and second, its summarizes the results by the median time for many repetitions, when the mean is what is needed. The median and mean can differ drastically, because just a few of the repetitions will include time for a garbage collection. These flaws can result in comparisons being reversed, with the expression that is actually faster looking slower in the output of microbenchmark.
Here’s an example of this, using R-3.0.2 on an Ubuntu Linux 12.04 system with an Intel X5680 processor. First some setup, defining a vector v to be operated on, a vector of indexes, m, for an initial part of v, and variables used to repeat expressions k times:
> library(microbenchmark) > set.seed(1) > v <- seq(0,1,length=100000) > m <- 1:42000 > k <- 1000 > n <- 1:k
We can now compare two ways of computing the same thing using microbenchmark:
> (m1a <- microbenchmark (u <- v[m]^2+v[m], u <- (v^2+v)[m],
+ times=k))
Unit: microseconds
expr min lq median uq max neval
u <- v[m]^2 + v[m] 549.607 570.2220 816.0170 902.6045 11381.40 1000
u <- (v^2 + v)[m] 472.859 682.3785 749.4225 971.9475 11715.08 1000
Looking at the median time, it seems that u <- (v^2+v)[m] is 816.0/749.4 = 1.089 times faster than u <- v[m]^2+v[m].
Alternatively, we could use system.time to see how long a “for” loop evaluating one of these expressions k times takes (k was set to 1000 and n was set to 1:k above):
> system.time(for (i in n) u <- v[m]^2+v[m]) user system elapsed 0.808 0.060 0.869 > system.time(for (i in n) u <- (v^2+v)[m]) user system elapsed 0.912 0.092 1.006
This gives the opposite result! From this output, it seems that u <- v[m]^2+v[m] is 1.006/0.869 = 1.158 times faster than u <- (v^2+v)[m]. Multiplying these factors, we see that the comparisons using microbenchmark and system.time differ by a factor of 1.089*1.158 = 1.26.
Maybe one could argue that a 26% error is often not that bad — I did fiddle the lengths of v and m for this example so that the error would actually make a difference in which expression seems faster — but the main reason people use microbenchmark is apparently that they think it is more accurate than system.time. So which gives the right answer here?
To find out, the first thing to do is to repeat the same commands again. As seen in the full output of this example, the repetition produces much the same results. (This isn’t always the case, though — appreciably different results in repetitions can occur when, for example, R decides to change the level of memory usage that triggers a garbage collection.)
We can also try the order="block" control option for microbenchmark. This tells it to do all 1000 repetitions of the first expression, and then all 1000 repetitions of the second expression, rather than randomly shuffling them, as is the default. The results aren’t much different:
> (m2a <- microbenchmark (u <- v[m]^2+v[m], u <- (v^2+v)[m],
+ times=k, control=list(order="block")))
Unit: microseconds
expr min lq median uq max neval
u <- v[m]^2 + v[m] 548.558 566.785 816.6940 830.7535 11706.04 1000
u <- (v^2 + v)[m] 471.602 543.331 743.3735 836.7810 12162.34 1000
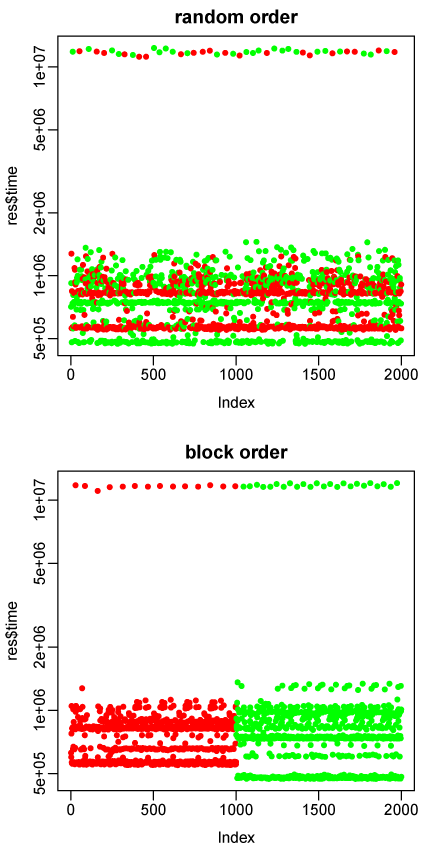
However, we can see what’s going on if we plot the individual times that microbenchmark records for the 2000 repetitions (1000 of each expression). Here are the plots for the second repetition with both random and block order:
The red dots are times for u <- v[m]^2+v[m], the green dots for u <- (v^2+v)[m]. Note the log scale for time.
Some of the repetitions take about 20 times longer than others. The long ones clearly must be for evaluations in which a memory allocation request triggers a full garbage collection. We can now see that the results using the median from microbenchmark are going to be wrong.
First, notice that with block ordering, 1000 evaluations of the first expression result in 14 full garbage collections, whereas 1000 evaluations of the second expression result in 24 full garbage collections. With random order, however, there are 20 full garbage collections during evaluations of the first expression, and 25 during evaluations of the second expression. Random ordering has obscured the fact that the second expression is responsible for almost twice as much garbage collection time as the first, presumably because it allocates larger vectors for intermediate results such as v^2 (versus v[m]^2).
But why, then, does block ordering give almost the same result as random ordering? Because we are looking at microbenchmark’s output of the median time to evaluate each expression. The median will be almost totally insensitive to the time taken by the small number (about 2%) of evaluations that trigger full garbage collections. So an expression whose evaluation requires more garbage collections will not be penalized for the time they take, even assuming that this time is correctly assigned to it (which happens only with block ordering).
This explanation suggests that using microbenchmark with block ordering, and then looking at the mean evaluation time for each expression would give a more accurate result, agreeing with the result found with system.time. And indeed it does in this example — in the two repetitions, system.time gives ratios of times for the two expressions of 1.158 and 1.136, while microbenchmark with block ordering gives ratios of mean times for the two expressions of 1.131 and 1.137. (These ratios of mean times can be computed from the data saved by microbenchmark, as seen in the R program I used, though the package provides no convenience function for doing so.)
So microbenchmark’s problem with this example can be fixed by using block ordering and taking the mean time for repetitions. But there’s no reason to think the time it then produces is any more accurate than that from system.time. It’s probably less accurate. The overhead of the “for” loop used with system.time is very small; in particular, since a change I suggested was incorporated in R-2.12.0, there is no memory allocation from the “for” loop iteration itself. The overhead of measuring the time for every repetition in microbenchmark is greater, and though microbenchmark attempts to subtract this overhead, one might wonder how well this works. Executing the time measurement code between each repetition will also disturb the memory cache and other aspects of processor state, which may affect the time to evaluate the expression.
More fundamentally, nanosecond timing precision is just not useful for this task, since to correctly account for garbage collection time, the number of repetitions must be large enough for many garbage collections to have occurred, which will lead to run times that are at least a substantial fraction of a second. Millisecond timing precision is good enough.
Furthermore, there’s no point in measuring the relative speeds of different expressions to more than about 5% accuracy, since recompiling the R interpreter after any slight change (even in code that is never executed) can change relative timings of different expressions by this much — presumably because changes in exact memory addresses affect the behaviour of memory caching, branch prediction, or other arcane processor tricks.
The data gathered by microbenchmark could be useful. For instance, it would be interesting to investigate the variation in evaluation time seen in the plots above beyond the obvious full garbage collections. Some is presumably due to less-than-full garbage collections; there could be other effects as well. But the microbenchmark package isn’t useful for the purpose for which it was designed.
Entry filed under: Computing, R Programming, Statistics, Statistics - Computing.
8 Comments Add your own
Leave a comment
Trackback this post | Subscribe to the comments via RSS Feed
1. Carl Witthoft | 2014-02-03 at 3:27 pm
Well, since the output of microbenchmark includes the max and min values, it’s dang easy to “guess” that there are occasional outliers. One can argue forever over whether median or mean is a more useful indicator of a statistical distribution, but that’s not the point here. At the very least, if two functions’ times as reported by microbenchmark are close, you should investigate to see how they compare with, e.g. 10 x and 0.1x the size of the input data.
2. Radford Neal | 2014-02-03 at 3:50 pm
The large values aren’t “outliers”. They happen when the evaluaton triggers a full garbage collection, which is an entirely expected event, the time for which has to be counted when assessing how much time an expression takes.
One can argue forever about anything. But often the argument for one side is correct, and the other is incorrect. The mean is the correct statistic to use here. The median is incorrect. You can see this by seeing which one gives a better match to the result of system.time. If you want to argue the other way, then what is your argument for why system.time is wrong?
Certainly, when the times for two expressions are close, one could suspect that the comparison might come out the other way if the situation were changed just a bit (eg, with different length vectors). So one should be caution about generalizing. I don’t see how looking at values that are off by 26% helps with this, however.
3. Carl Witthoft | 2014-02-03 at 3:57 pm
Well, what’s the statistical spread for system.time() over 100 trials? I know from experience it can vary by 10% (at least) over 3 or 4 runs.
4. Radford Neal | 2014-02-03 at 4:02 pm
Yes, the result of system.time can vary, for many reasons. Some are external, such as load from other users or system processes. Some are due to hardware (some processors adjust their clock frequency based on whether the processor is getting too hot). Some are internal to R, such as automatic adjustment of the level of memory usage that triggers a garbage collection.
The results from microbenchmark will vary for the same reasons.
5. Hadley Wickham (@hadleywickham) | 2014-02-03 at 7:47 pm
I think the meat of your argument is that you think microbenchmark should include the cost of gc, while it seems like the author does not. I can see arguments either way. Since it’s not possible to unambiguously allocate the costs of gc to an individual expression (unless you want to do gctorture = TRUE), ignoring the cost of gc (as the median does) seems to me to be a reasonable approach. The other approach, to run the expression a sufficiently large number of times to get an estimate of average gc cost also seems reasonable.
To me, the advantage of ignoring gc and timing individual expressions is better statistics. You have better experimental design since you’re randomising the order of the experiments, and you get a measure of variability. system.time() gives no information about variability, so if you want to make a statistical comparison you’ll need to run it multiple times. If you have to run it multiple times, you might as well use microbenchmark (which of course you can use with a loop).
But perhaps you mostly argue against microbenchmarking in general (as it is rarely useful for understanding the performance of real code), not the microbenchmark package specifically.
6. Radford Neal | 2014-02-03 at 9:38 pm
Microbenchmarking a small bit of code certainly could be misleading, since, for example, the parts of the interpreter used by the small bit of code tested will tend to stay in cache, whereas they might not if many other parts of the interpreter were also being activated by R code. That issue is not the focus of my post, however. I’m assuming we want to time the little bit of R code on its own, and get the correct answer within that context.
Ignoring garbage collection time amounts to pretending that memory allocation takes less time than it really does. That makes little sense to me. I don’t think allocating the costs of gc to individual expressions is entirely ambiguous – though there are some complications, the responsibility for garbage collection roughly tracks the amount of memory the expression allocates. And in any case, using the median time from microbenchmark isn’t going to reliably exclude all time spent in garbage collection (including partial garbage collections). If you want to separate out gc time, you can use gc.time (though it was broken until recently).
Looking at the median time isn’t a good idea even apart from garbage collections. Suppose that cache performance varies with the exact memory locations used to store objects. If in 1000 repetitions, 550 get lucky in memory locations used, and 450 are unlucky and take twice the time, do we really want to use the median, essentially pretending that we’ll always be lucky?
I think we want the mean time. And if we want an indication of uncertainty, a standard error for the estimate of the mean time is what would be appropriate – though it will be only a lower bound on true uncertainty, since there are many factors that affect the mean time that won’t have varied during a short test. We don’t, for most purposes, want to know the variability over individual repetitions, since we already know that this will be huge, with a long tail due to occassional garbage collections.
7. Hadley Wickham (@hadleywickham) | 2014-02-04 at 9:32 am
A single number summary of a distribution is never going to be perfect. For example, there’s no way to accurately summarise a bimodal distribution with only one number. You can argue that the mean makes more sense in your example. But there are plenty of other cases where you can argue the median makes more sense (i.e. if you have one large outlier).
Having the distribution of times gives you the flexibility to choose your summary and estimate its variability after the fact. Using system.time() only gives you one summary statistic and no ability to calibrate uncertainty. If you want to compare two operations using system.time() you’ll need to think about your experimental design to make sure external system factors aren’t influencing the results.
It feels to me like the framework provided by microbenchmark is more likely to give useful results. Certainly there are improvements that can be made. Perhaps it should default to a block random design where each expression is run n/m times, randomly interleaved. Perhaps the default summary should display a mean with confidence interval. These changes would not be difficult to make, and I’m sure are things that Olaf will consider.
8. Arun | 2014-02-27 at 4:24 am
@Hadley, I think outliers can be tackled, for example, by calculating `winsorised` mean. It would provide better timings that reflect the effect due to cache misses quite nicely still. But with just ‘median’ it might not be possible to measure cache effects at all in certain scenarios (as the one in Radford’s commentl.
I’ve nothing against `microbenchmark` package, it’s great. But seems like it certainly could be improved.