Evaluation of NUTS — more comments on the paper by Hoffman and Gelman
2012-01-27 at 4:31 pm 14 comments
Here is my second post on the paper by Matthew Hoffman and Andrew Gelman on “The No-U-Turn Sampler: Adaptively Setting Path Lengths in Hamiltonian Monte Carlo”, available from arxiv.org. In my first post, I discussed how well the two main innovations in this “NUTS'” method — ending a trajectory when a “U-Turn” is encountered, and adaptively setting the stepsize — can be expected to work. In this post, I will discuss the empirical evaluations in the NUTS paper, and report on an evaluation of my own, made possible by the authors having kindly made available the NUTS software, concluding that the paper’s claims for NUTS are somewhat overstated. The issues I discuss are also of more general interest for other evaluations of HMC.
The scripts used for the empirical evaluations in the NUTS paper are not included with the NUTS software, so in some respects I’m not certain what was done, but the description in the paper is fairly complete.
The foundation for any empirical assessment of MCMC methods is some way of estimating how good the MCMC estimates are, which could be the autocorrelation time, or the variance of the estimate, or the effective sample size (ESS, the equivalent number of independent points). The NUTS paper uses autocorrelation time, from which they compute the effective sample size. They estimate autocorrelations using means and variances found from a very long run, not from the run being evaluated, which avoids one possible source of drastically wrong results in such evaluations. (For the zero-mean multivariate normal example I discuss below, they presumably plugged in the true mean of zero and the true variances, rather than use a long run.) As their measure of overall performance for a sampler, they used its worst performance (in terms of autocorrelation time or ESS) over all variables (looking at both mean and variance estimates for these variables). Focusing on the worst estimate seems reasonable in a way, but looking at only this one number could be misleading if the estimate that is worst is different for different samplers.
One aspect of their evaluation (as described in the paper) is incorrect. In the Appendix, they say that they estimate autocorrelation time by summing autocorrelation estimates at all lags up to the first lag at which the autocorrelation estimate is less than 0.05. Since the true autocorrelation can sometimes be negative, this is not valid, in general — it would be for some simple random-walk Metropolis methods, for which autocorrelations are all positive, but not for HMC, for which negative autocorrelations are quite possible. However, for reversible methods (which includes HMC), the sums of consecutive pairs of autocorrelations are always positive. This provides the basis for a similar approach that is valid. See here for a review and assessment of this and other approaches to estimating autocorrelation time.
Assuming that this is an error in the actual code (not just the paper), it could affect the empirical results quite a bit, but I’ll have to ignore this possibility in the remaining discussion.
The first distribution that Hoffman and Gelman test NUTS on is a 250-dimensional multivariate normal distribution with mean zero and a covariance matrix randomly drawn from an inverse Wishart distribution. They claim that NUTS performs better on this distribution than HMC (by a factor of about three) even when HMC is optimally tuned. This seems strange, since NUTS is just an automated way of tuning HMC. As I’ll show below, this claim is indeed likely to be due to deficiencies in the experimental methodology.
Due to rotational invariance, performance of HMC on a multivariate normal is determined by the set of eigenvalues of the covariance matrix, with the inherent difficulty of the problem being proportional to the square root of the ratio of the largest eigenvalue to the smallest eigenvalue. This is the ratio of the standard deviation in the least confined direction to the standard deviation in the most confined direction, and should be roughly the number of leapfrog steps in a trajectory of optimal length, using the optimal stepsize. (Performance by Hoffman and Gelman’s metric isn’t quite rotationally symmetric, however, since they look only at autocorrelation times for the 250 variables, not all linear combinations of them.) The evaluation of HMC and NUTS on this distribution would be much more informative if the eigenvalues of the covariance matrix were reported, and if the number of leapfrog steps in HMC trajectories (not just the product of stepsize and number of steps) were reported.
I’ll return below to the multivariate normal example, but first I’ll mention the other distributions on which HMC and NUTS are evaluated in the paper. Two of these other distributions are posterior distributions for logistic regression models. The results on these distributions are more what one might expect — NUTS performs about as well as HMC with optimal tuning. One slightly strange phenomenon is visible in the results presented in Figure 6, however. For both logistic regression problems, the performance of HMC drops drastically as the trajectory length is increased from the optimal value to a value larger by a factor of 1.5. I would expect a more gradual decline in efficiency. It would be nice to know why this occurs.
The last distribution they evaluate NUTS and HMC on is the posterior distribution for a stochastic volatility model. Here again, they claim that NUTS performs better than HMC, even when the latter is optimally tuned. I’m not sure what is going on here, but one thing I notice in the lower left plot in Figure 6 is that the performance of NUTS seems to be bimodal — some of the runs are assessed as being three times better than the best HMC runs, but others are assessed as being worse. All I can think of to explain this bimodality is that the adaptive stepsize selection might be reaching either of two stable values. The advantage of some NUTS runs over the best HMC runs might be explained by the same factors that lead to this result for the multivariate normal.
Since the actual multivariate normal distribution used in the paper isn’t reported, I’ll look at performance of NUTS and HMC on another multivariate normal, the same 30-dimensional multivariate normal distribution that I used in my previous post, in which the 30 components are independent, with standard deviations of 110, 100, 1.1, 1.0, and 26 values equally spaced between 8 and 16. This may in any case be a better demonstration example than one in which the standard deviations are chosen randomly.
I used the NUTS software for these experiments, with automatic stepsize adaptation for both HMC and NUTS (the adaptation always converged to a reasonable value of around 1.63 for HMC and around 1.73 for NUTS). I did 10,000 burn-in iterations, followed by 250,000 sampling iterations, which I divided into 400 batches of size 625. I estimated the mean of each variable and the second moment of each variable (equal to the variance, since the true mean is zero) for each of the 400 batches, and looked at the variance of these batch estimates as a measure of the performance of the sampler, not accounting for computation time yet. I then multiplied the variances of these batch estimates by the number of gradient evaluations for the whole run to get figures of merit that account for computation time. I used this Matlab script for these experiments (and then moved the results to R for plotting).
Here are the results for estimating the means of each of the 30 variables (numbered 1 to 30 by decreasing standard deviation), for HMC with trajectory lengths of 100, 170, and 200 leapfrog steps (and hence trajectory lengths of about 163, 277, and 326 time units since the stepsize is about 1.63):

The left plot gives the variances of the estimates, not accounting for computation time; the right plot adjusts for computation time (gradient evaluations). Smaller is better in both plots. Note that the vertical scale is logarithmic, with horizontal lines at each power of ten.
These results look rather peculiar! Notice how the variances of the mean estimates for variables 3 to 28, with standard deviations declining from 16 to 8, don’t decline as one might expect, but rather go up and down, with peaks at positions that change with the trajectory length.
This behaviour is due to a phenomenon discussed in Section 3.2 of my review of Hamiltonian Monte Carlo. Hamiltonian dynamics for this simple distribution without dependencies between variables will be periodic for each variable. If the period for a variable happens to match (or be near) the trajectory length, the end-point of a trajectory will be close to the start point, with the result that sampling for that variable is very poor. (A related problem happens when the trajectory length is half the period.) Note that this problem can arise for any multivariate normal distribution, with the poorly-explored directions being the eigenvectors of the covariance matrix, not necessarily the individual variables.
The cure for this is to randomly vary the trajectory length (randomly varying the stepsize, or the number of leapfrog steps, or both) over some moderate range. I modified the hmc_da function in the NUTS software to do this, changing the computation of the number of leapfrog steps from
L = max(1, round(lambda / epsilon));
to
L = max(1, round((0.9 + rand/5) * lambda / epsilon));
which has the effect of multiplying the number of leapfrog steps by a random factor uniformly distributed between 0.9 and 1.1.
Here are the results after this modification:

The variances of the mean estimates still go up and down a bit (maybe a wider random range would be better), but much less than before.
So we see that one can get a misleadingly-bad impression of the performance of HMC on a multivariate normal distribution from experiments in which the trajectory length is not varied randomly . On less simple distributions, the exact periodicities that happen with a multivariate normal may not be present, but there can be approximate periodicities, which might still happen to nearly match a fixed trajectory length, with a consequent bad effect on performance. This is why randomly varying the trajectory length is a recommended part of standard HMC methodology. (However, this is unnecessary with “windowed” HMC, as long as the window size is a non-negligible fraction of the trajectory length.)
So, how does NUTS perform on this distribution? Here are the results, together, for comparison, with the variances of mean estimates from an equal-size sample of points drawn independently (omitted for the computational cost plot, where it would not be meaningful):
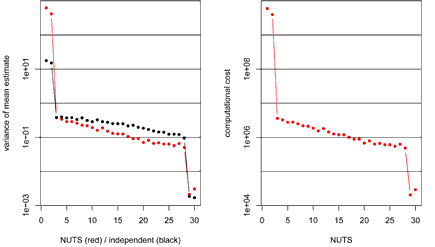
For the first two variables, with the largest standard deviations, the variances of the NUTS estimates of the mean are about 30 times bigger than the variances when the sampled points are independent. Compared to HMC with random trajectory length with median 200, these NUTS estimates have a variance that is about 25 times bigger. After adjusting for computational cost, the NUTS estimates of the means of these two variables are 7 times less efficient than the HMC estimates.
NUTS looks much better for the other variables, with smaller standard deviations. Indeed, the NUTS estimates are often better than those obtained with independent sampling! This is possible because NUTS (and HMC) can produce negative autocorrelations. However, for negative correlations to show up with these variables, NUTS must often be using trajectories that are much shorter than are needed for efficient exploration of the large-standard-deviation variables, which of course is consistent with it estimating the means of those variables inefficiently.
Here are plots showing the performance of HMC with random trajectory length and of NUTS when estimating the variances (rather than the means) of the 30 variables:


HMC is about a factor of two more efficient than NUTS at estimating the variances of the two variables with the largest standard deviations, but is less efficient at estimating the variances of the other variables.
One might think that these results show that there’s a tradeoff — HMC better for variables with large standard deviation, NUTS better for variables with small standard deviation. But I think this isn’t really so. First, it is often the absolute magnitude of the variance of estimates that is important, and in that respect HMC is clearly better. (Consider, for instance, estimating regression coefficients for covariates that have been standardized.) When that’s not so — when you need precise estimates of the means of variables that have small standard deviations — it’s easy to alternate HMC updates designed to sample well for the variables with large standard deviations with other, much cheaper, updates designed to sample well for the variables with small standard deviations (eg, univariate slice sampling updates, or just HMC with a smaller trajectory length). Using the windowed HMC method would also help, since rejected transitions in windowed HMC will usually move a small distance, helping with the sampling for variables with small standard deviations. (Note: as mentioned before, due to the rotational invariance of HMC, all this discussion applies also when the “variables” are actually various directions in the space, that aren’t necessarily the coordinate axes being used.)
One conclusion I draw from this evaluation is that the better performance of NUTS over HMC (optimally tuned) shown in the evaluations in the NUTS paper is likely an artifact of not randomizing the HMC trajectory length, and of evaluating performance by the worst effective sample size for any variable (not accounting for some variables being easy to handle by other methods). Another conclusion is that the premature U-Turn problem that I discussed in my first post on NUTS can be a problem in practice, since otherwise NUTS would estimate the means and variances of the large-standard-deviation variables in this example better. As I discussed there, I think there is scope for improving the way that NUTS decides when to stop a trajectory, and thereby make it a more generally-useful sampling method.
Entry filed under: Monte Carlo Methods, Statistics, Statistics - Computing.
1. Bob Carpenter | 2012-01-30 at 1:31 pm
NUTS isn’t “just an automated way of tuning HMC” — the number of leapfrog steps can vary from sample to sample.
For our Stan implementation of HMC and NUTS, we were planning to allow at least HMC to have randomly varying step sizes according to your advice in your MCMC Handbook paper on HMC.
We have also been thinking about ways to block subsets of the variables for more flexibility, but that’s not going to make it into the first official release.
2. Radford Neal | 2012-01-30 at 2:09 pm
Yes, the number of steps that NUTS takes can vary from one iteration to the next, but is that good or bad? To say that it’s more than an automated way of tuning HMC one would need to demonstrate that this variation is useful, and in particular that it has a dependence of the current point that is useful (since otherwise it’s just a way of tuning a distribution for the number of steps rather than a single value). For a multivariate normal distribution, I don’t think there is any useful way of varying the number of steps according to the current position.
Being able to block subsets of variables would certainly be helpful, possibly even essential. In particular, I’m skeptical that updating variance hyperparameters with HMC or NUTS is going to work well, at least without some non-obvious trick, because of the drastic changes in probability density that will accompany changes in a variance hyperparameter that controls many lower-level parameters.
3. Jascha Sohl-Dickstein | 2012-02-12 at 8:16 pm
Both the paper, and this commentary, have been very interesting reads. Thank you!
My understanding of Hoffman’s and Gelman’s algorithm is that the number of steps varies as a function of the starting location in both position and momentum space, not just position space. Even for a multivariate normal distribution, I would think varying the number of steps depending on whether the momentum vector is pointing in a low or high eigenvalue direction could be extremely useful. Their algorithm should tend to take more steps when the momentum vector is pointing in a low curvature direction, and at least qualitatively this sounds like desirable behavior.
4. Longhai Li | 2012-02-23 at 12:25 am
This is an interesting example. Just a question related to this example that bugs me: if one can transform a target distribution into something like the example you show, do you think there is still any benefit of using HMC at all? It seems to me that applying slice sampling one-by-one should work as well as HMC, without worrying about the stepsize and length of trajectory. Slice sampling asks for choice of stepsize, but slice sampling can quickly cut off the overly long slice and so a overly long stepsize should not be worried as much as the tuning of HMC. It seems to me that this is an advantage of using slice sampling for no-correlated distribution. Also, I guess for most problems, we are not totally blind to the widths of the univariate distributions.
5. Radford Neal | 2012-02-23 at 8:42 am
Certainly in low-to moderate dimensions there is no need to use HMC to sample from a multivariate Gaussian distribution – finding the Cholesky decomposition of the covariance matrix lets you sample independently in n-cubed time for setup and n-squared time for each point after that. In higher dimensions this takes too long, but there are other specialized methods that might be better than HMC, though I think in some cases HMC might still be the best choice. (Univariate slice sampling would work well only if you have transformed to make variables independent, which takes the same time as using the Cholesky decomposition.)
Looking at a multivariate Gausian distribution is mostly a way of getting insight into what happens with less siimple distributions. That’s why it’s important to avoid the effects of periodicities, which are not a good guide to what will happen with most distributions.
6. Nigel Goodwin | 2012-05-18 at 11:39 pm
I saw a comment on the previous post about using second derivative information, and using a weighting matrix. There has been a recent paper (Zhang and Sutton, Edinburgh) on this about using a BFGS approximation – wouldn’t this alleviate most of the difficulties, and is not too costly to perform (in my applications the function and gradient evaluation is quite expensive, a 100 dimensional back substitution for the second derivatives would not figure on the radar, but the function has a very complex behaviour).
7. jsalvati | 2013-01-06 at 3:55 am
I was initially excited by Zhang’s paper, but experience with it was that if the number of stored samples was significantly less than the number of dimensions of the distribution, the sampler got ‘stuck’ in a subspace of the true distribution. It would only slowly move orthogonal to that distribution.
I suppose it’s possible I misimplemented the algorithm, though. If you’ve had more success, I’d love to hear about it.
8. Nigel Goodwin | 2013-01-06 at 10:58 am
I have done a lot of work on this BFGS method and NUTS. Long story short, if the hessian is reasonably constant over the space, it can give excellent benefits. E.g. tests on a highly correlated Gausian in high dimensions was very impressive. But in more realistic functions, such as Harrio banana functions, the Hessian approximation is not good, and behaviour can be worse than no Hessian.
I’m afraid the holy grail has not been found. We need a hessian which is local along leapfrog steps, rather than global, we need a hessian which can be efficiently calculated and/or adapted with efficient leapfrogs, and we need to be able jump modes. No published method I have seen satisfies these criteria.
Also of course it has to satisfy detailed balance etc. I tried various methods which did not satisfy detailed balance but gave good local hessians, but the results were (not surprisingly) unreliable.
I have had lengthy discussions with Hoffman and Zhang, and I think we all came to similar conclusions.
My current approach is to use a mix of different methods – NUTS and DREAM and RWM, each has strengths and weaknesses.
9. jsalvatier | 2013-01-06 at 2:48 pm
Thanks for the update, very interesting.
Do you have any thoughts about “BAYESIAN ADAPTIVELY UPDATED HAMILTONIAN MONTE CARLO WITH AN APPLICATION TO HIGH-DIMENSIONAL BEKK GARCH MODELS”?
Click to access wp46_12.pdf
or “Lagrangian Dynamical Monte Carlo” ?
http://arxiv.org/abs/1211.3759
10. Nigel Goodwin | 2013-01-06 at 5:07 pm
I was kind of aware of the Lagrangian approach, it looks promising, but I won’t be implementing and testing it for some time – I have other priorities. It is difficult enough for me to calculate derivatives of my function, let alone second derivatives. My function is based on linear regression, radial basis functions, and Matern functions, and is used to calculate an estimate and a variance, and the variance contributes to the objective. Not so simple.
As I come from an optimisation background, BFGS is very appealing, but not so simple to see how it can be used in an MCMC scheme.
I do all this for optimisation of complex engineering designs.
The Burda and Maheu paper looks interesting, but I have been advised by others (who are of good repute) that it may not be reversible, so I would treat it with caution.
DREAM and its variants can be very useful. It is a nice idea.
11. Yichuan Zhang | 2013-06-10 at 12:04 pm
The initial diagonal Hessian approximation is important in practice to prevent moving in a subspace. We should say something about it in the paper. Unfortunately, that means you will have one more parameter (vector) to tune, the diagonal hessian approximate H_0.
12. Nigel Goodwin | 2012-05-18 at 11:55 pm
ps I am currently implementing an integrated NUTS/BFGS solution to see what happens, but there will be no published paper or detailed evaluation as I am not an academic but work for private industry. I first started using BFGS way back in 1978.
13. Nigel Goodwin | 2012-05-27 at 5:09 pm
I’ve been doing my own investigation of NUTS.
Caveat – I have converted from Matlab to Java, so I may have introduced some bugs, but I can’t find any…without a Matlab environment, it is difficult to trace through and identify any differences.
One thing to be careful of is statements like:
if (rand() < nprime2 / (nprime + nprime2))
where you have to understand how Matlab treats divide by zero.
My initial conclusion is that NUTS does not work at all well on the example problem on the web site, which is 2D and I used R to find the eigenvalues and vectors. It works fine on a multivariate with unit variances. My java implementation does not come close to the analytical solution, whereas my implementations of HMCMC are good. So my first assumption is that this is a Java bug, but would be interested if anybody has tested this function and their results. Is it correct that sometimes (often?) nprime and nprime2 are both zero?
Not sure why I am posting this on this blog rather than the NUTs authors – let me take a look at their blogs.
function [logp grad] = correlated_normal(theta)
A = [50.251256, -24.874372; -24.874372, 12.562814];
grad = -theta * A;
logp = 0.5 * grad * theta';
Of course I may be talking to a wall…..
14. Nigel Goodwin | 2012-05-27 at 9:09 pm
Good news – I downloaded Octave and ran the example, it does what it should according to analytical solution, so it is some bug in my java conversion, which should now be easy to sort out.
Isn’t Octave slow, compared to Java!